library(igraph)
library(ADAPTSNA)7 Cleaning Network Data - Subgraphs
You may want to create subgraphs of the network that you have. There are two basic ways that you can think about this. You may be interested in a specific group of people and how they relate to each other, or you may be interested in a specific person and find out who they are connected to.
| LEARNING ELEMENTS - Data Discoveries |
|---|
|
First, we start by bringing in the data and cleaning out the self loops. This new dataset is of some Grime musicians from Spotify. The nodes are the artists and the ties represent collaborations between the artists.
grime_edge_list <- load_data("GRIME_2008_Edge.csv", header = TRUE)
grime_08 <- graph_from_data_frame(d= grime_edge_list, directed = TRUE)grime_08_clean <- delete_edges(grime_08, E(grime_08)[which_loop(grime_08)])7.1 Specific Subgraphs
First, let’s talk asume you need a subgraph to see a specific set of people and how/whether they are connected. You may have a list of individual nodes that you are interested in and you want to see how they related to each other. You can do this by creating a vector with the names of those nodes, then use the subgraph function().
Why might you want to do this? There could be a highly prominent individual, or group of individuals in the network and you might want to see how these individuals are connected. Here, the individuals, Wiley, Jammer, Flowdan, and Ice Kid, are some of the older generation grime artists. In this network, taken from 2008, it might be interesting to see how/if these individuals are connected.
sub_people <- c('Wiley', 'Jammer', 'Flowdan', 'Ice Kid')
sub_net <- subgraph(grime_08_clean, sub_people)
par(mar = c(0,0,0,0))
plot(sub_net)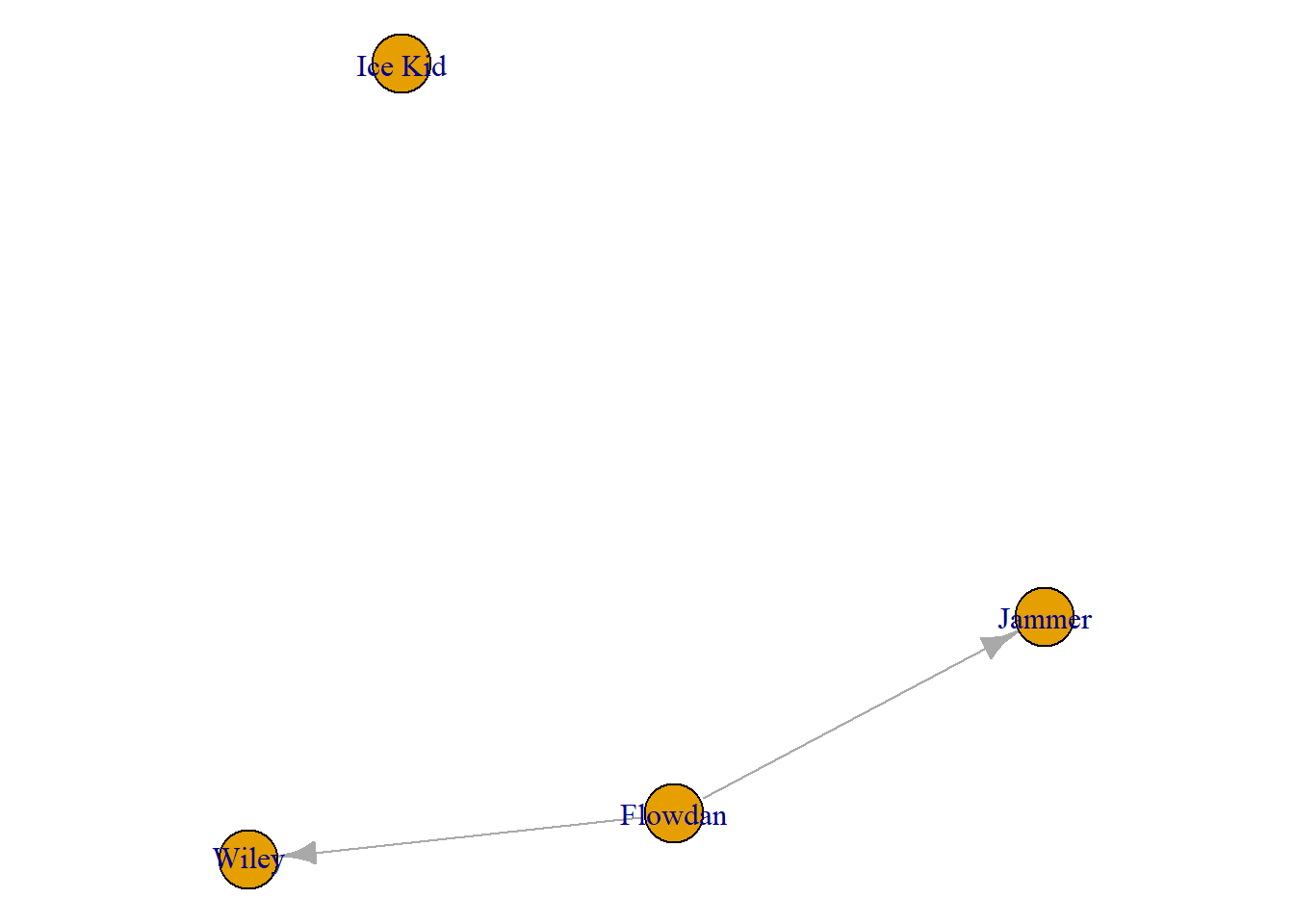
7.2 Ego Graphs
Next, you may want to see ego networks from those in your network. In other words, smaller networks showing only the connections of each individual artist. To do this, you can use the make_ego_graph() argument. This creates a list of ego graphs from your entire network. Note, the order = 1 argument refers to the number of steps away from the ego (focal node). Since mine is set to 1, this only captures the ego’s immediate neighbours (i.e. only those directly connected to ego).
ego_graphs <- make_ego_graph(grime_08_clean, order = 1)
head(ego_graphs)[[1]]
IGRAPH 7307b90 DN-- 2 1 --
+ attr: name (v/c), collab_weight (e/n)
+ edge from 7307b90 (vertex names):
[1] Asher D->Wiley
[[2]]
IGRAPH 7307ba9 DN-- 1 0 --
+ attr: name (v/c), collab_weight (e/n)
+ edges from 7307ba9 (vertex names):
[[3]]
IGRAPH 7307bb5 DN-- 1 0 --
+ attr: name (v/c), collab_weight (e/n)
+ edges from 7307bb5 (vertex names):
[[4]]
IGRAPH 7307bbf DN-- 2 1 --
+ attr: name (v/c), collab_weight (e/n)
+ edge from 7307bbf (vertex names):
[1] Scorcher->Wiley
[[5]]
IGRAPH 7307bca DN-- 3 2 --
+ attr: name (v/c), collab_weight (e/n)
+ edges from 7307bca (vertex names):
[1] Bless Beats->Wiley Bless Beats->Roll Deep
[[6]]
IGRAPH 7307bd6 DN-- 3 2 --
+ attr: name (v/c), collab_weight (e/n)
+ edges from 7307bd6 (vertex names):
[1] Flowdan->Wiley Flowdan->JammerYou can also specify exactly which node’s network you want to see. Let’s say there was a person of interest in your network that you specifically want to see. To do this, you can do the following using the node’s name to single them out.
This chunk returns a list of edges connected to Wiley (the name of my node of interest).
E(grime_08_clean)[[.inc('Wiley')]]+ 8/28 edges from 72f5cd5 (vertex names):
tail head tid hid collab_weight
1 Asher D Wiley 1 29 1
2 Scorcher Wiley 4 29 4
3 Bless Beats Wiley 5 29 1
4 Flowdan Wiley 6 29 3
5 Tinchy Stryder Wiley 7 29 2
6 Frisco Wiley 8 29 1
7 Kano Wiley 9 29 1
27 Wiley Lauren Mason 29 39 1I can also plot these. To do so, I make an object with the name ‘Wiley’ and then make an ego graph based on that name only. The [[1]] simply tells R to get only the first one in the list that make_ego_graph() creates. In this case, Wiley. Using the “order = 1” option, you are selecting to gather Wiley’s immediate neighbours (known as a first order ego network).
Wiley <- "Wiley"
ego_wiley <- make_ego_graph(grime_08_clean, order = 1, nodes = Wiley)[[1]]
par(mar = c(0,0,0,0))
plot(ego_wiley)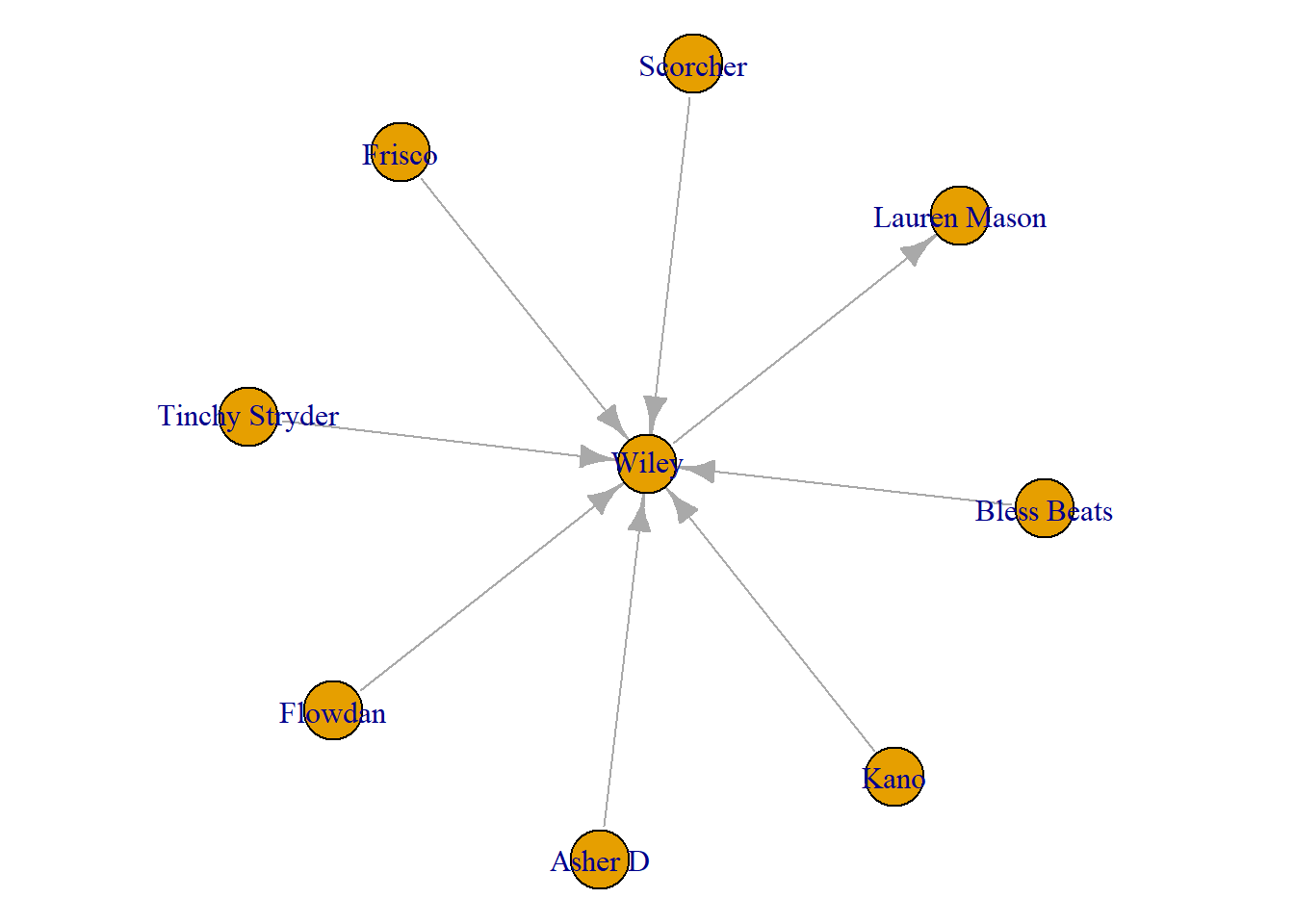
The second order ego network includes the connections of Wiley’s neighbours. This is useful to see whether/how Wiley’s connections are also collaborating.
second_order_wiley <- make_ego_graph(grime_08_clean, order = 2, nodes = Wiley)[[1]]
par(mar = c(0,0,0,0))
plot(second_order_wiley)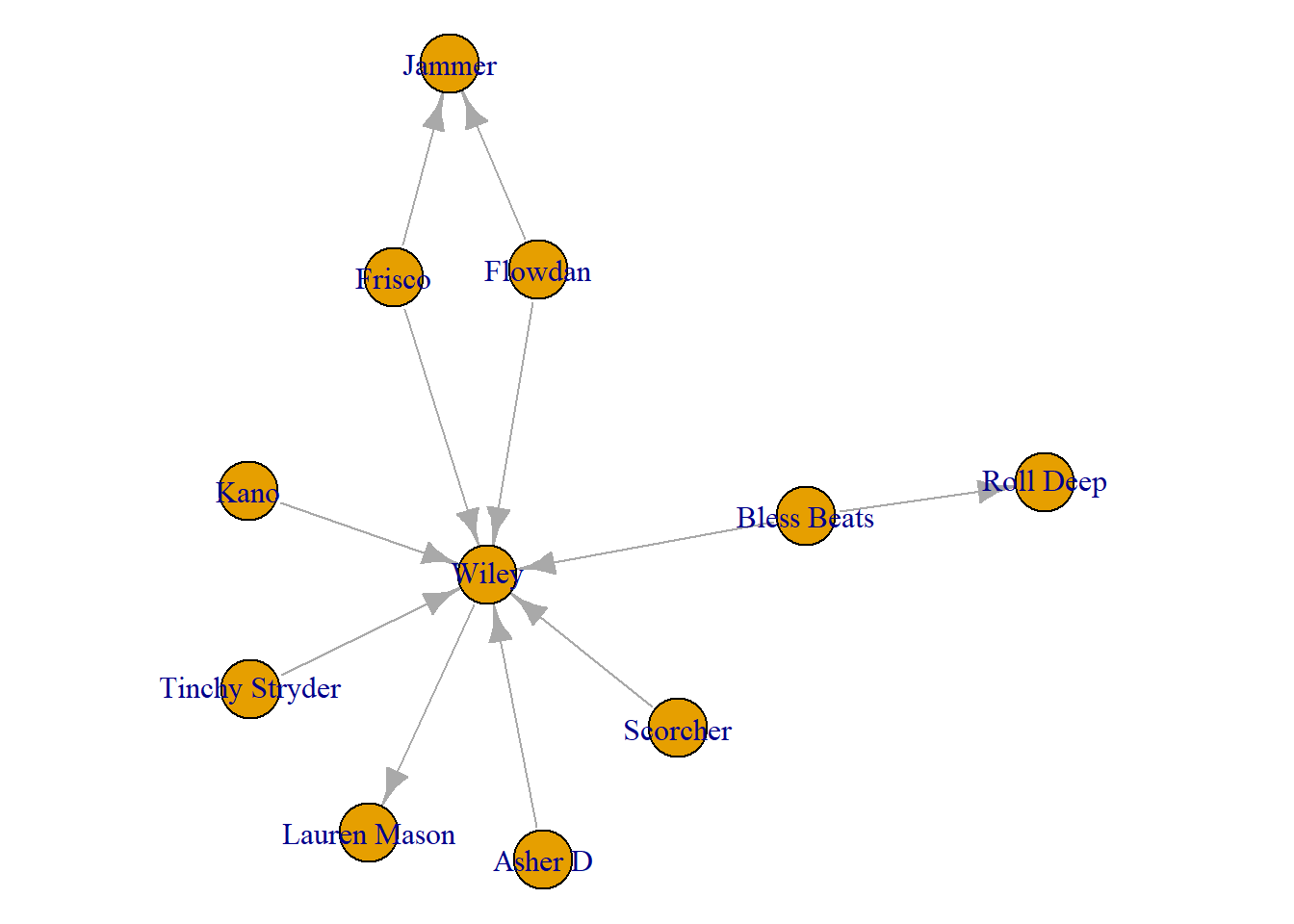
Pro tip: If you are working with ego networks like this, especially when you get passed the first order network (including friends of friends) it is good practice to do something to differentiate the ego from their neighbours. This way, someone who is looking at the graph can clearly identify who is the ego and who are the neighbours. One simple way it to change their colour.
Don’t get too caught up in this code below. We will cover a lot more of this in future chapters (see Chapter 11. What we do here is create a node characteristic called ‘ego’. What this characeristic does is assign colours to every node in the network. If the name of that node is “Wiley” then the colour is red, otherwise it is white. The next chunk changes the parameters of the plot so we can see it a bit easier. Then, using the vertex.color option of the plot() function, we change the colour of the visualisation to reflect the red and white that we just added.
V(second_order_wiley)$ego <- ifelse(V(second_order_wiley)$name %in% c("Wiley"), "red", "white")
par(mar = c(0,0,3,0))
plot(second_order_wiley, vertex.color = V(second_order_wiley)$ego, main = "Wiley's Second Order Ego Network")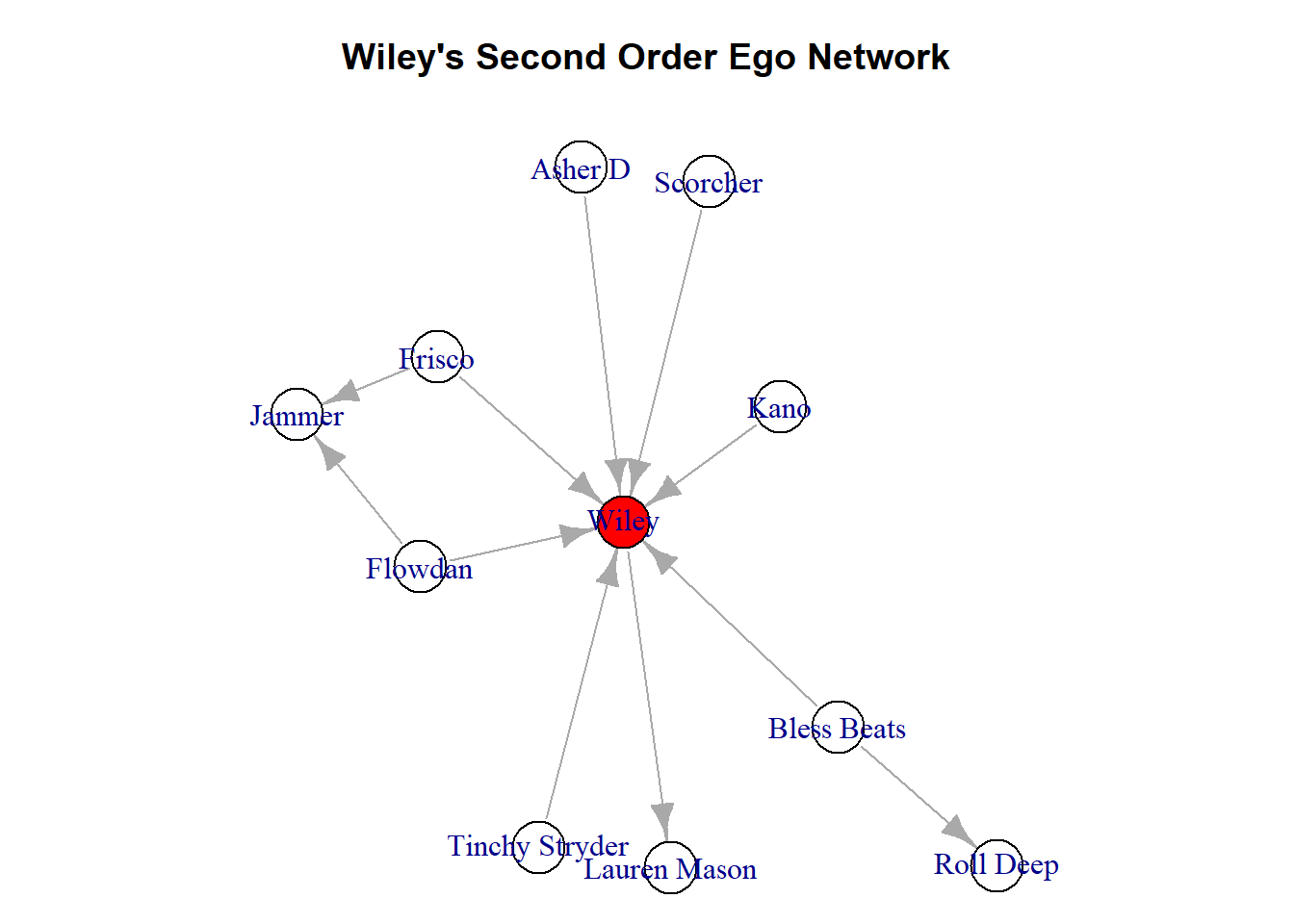
Finally, one other way to can subset a network is by a set parameter you may have. For example, you may want to see a network of frequent collaborators (more than 1 collab).
The following returns a vector with collaborators who work together more than once.
frequent_collabors <- E(grime_08_clean)[[collab_weight > 1]]
frequent_collabors+ 8/28 edges from 72f5cd5 (vertex names):
tail head tid hid collab_weight
2 Scorcher Wiley 4 29 4
4 Flowdan Wiley 6 29 3
5 Tinchy Stryder Wiley 7 29 2
8 Blacks Jammer 12 35 4
9 Badness Jammer 13 35 5
11 Tempa T Jammer 15 35 2
14 Skepta Jammer 17 35 5
16 Frisco Jammer 8 35 3You can then turn this vector of edges into a igraph object to plot.
frequent_collabors_graph <- induced_subgraph(grime_08_clean, vids = unique(c(ends(grime_08_clean, frequent_collabors)[, 1], ends(grime_08_clean, frequent_collabors)[, 2])))
plot(frequent_collabors_graph)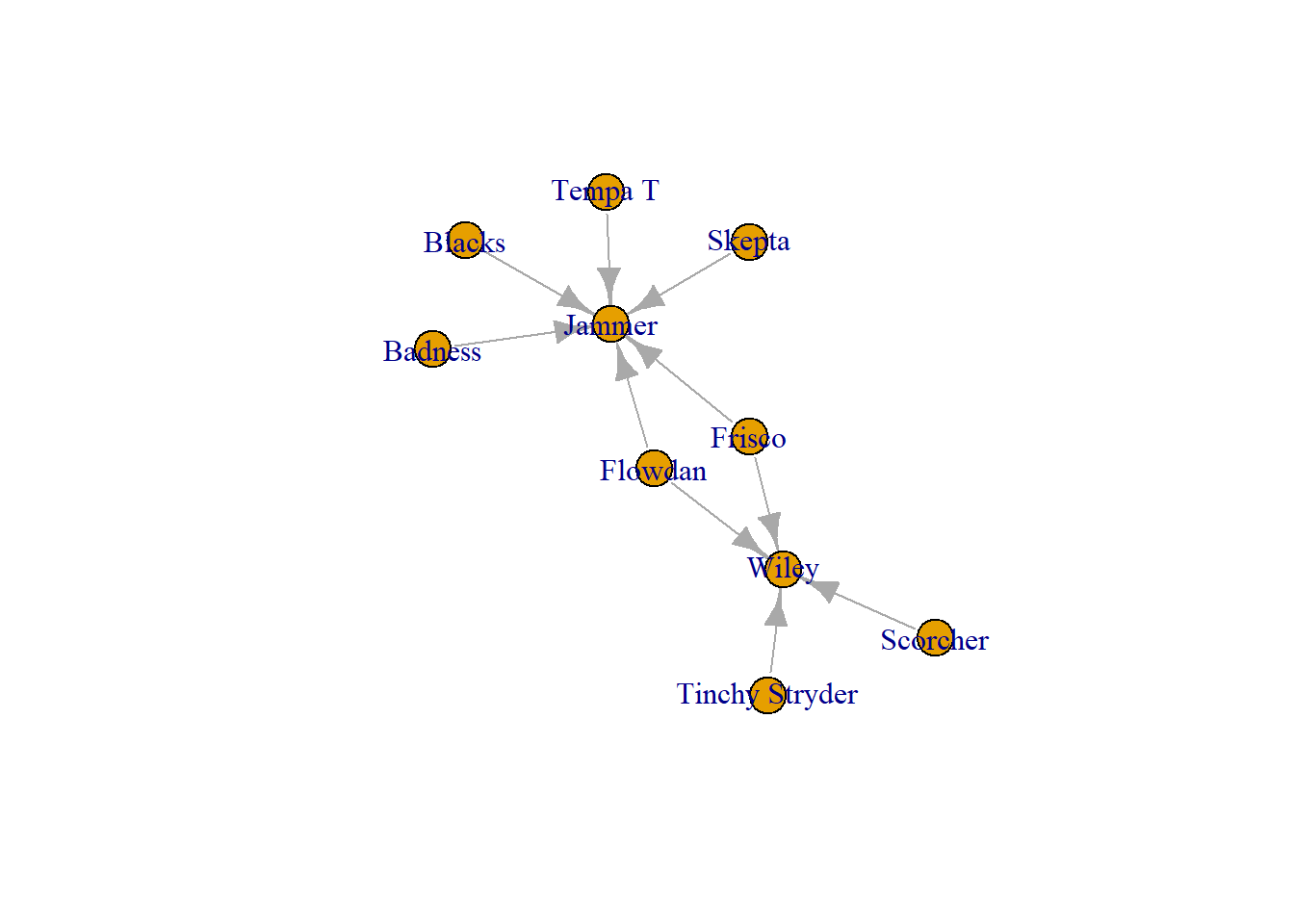
7.3 Summary
Here we have discussed another method for cleaning network data, taking a subgraph. This is a simple cleaning or transformation tool that allows you to study a subset of your data. We have covered how to take a specific subset based on the names of particular nodes of interest. Alternatively, we can we can create ego networks.
